

Enabling clinicians and patients to make data-driven decisions with genomics
Nick Lench, Genomics Expert and Co-Founder of Congenica, discusses the challenges holding back the large scale implementation of genomics and how automation will usher in a new era of personalised medicine
Genomics offers the tantalising promise of a future where patients can use a bounty of genetic and genomic information to make truly data-driven decisions about their own health and lifestyle. A huge amount of progress has been made towards this goal. Already, the cost of sequencing genomes has dropped from more than $1bn to under $1000. A number of companies are currently locked in a race to achieve ‘the $100 genome’.
However many challenges remain between now and this new era of healthcare.
To better understand these challenges, and how to overcome them, we spoke with Nick Lench, a genetics expert with decades of experience leading genomics teams and developing automated solutions for the genomics space. Today, he is Co-Founder of Congenica, the world’s leading clinical decision support platform for accelerated interpretation of genome sequence data .
Nick talked to us about how far genomics has come, how much room there is to improve the way labs manage genomic testing, and how technology – specifically automation – is necessary to overcome the challenges of scaling genomic testing.

“When I was undertaking my PhD between 1985-1990, if we managed to sequence 200 base pairs of DNA in a single run we’d get very excited,” he tells us. “So, if you’d told me back then we’d be able to conduct whole genome sequencing within my lifetime, I might not have believed you. The technology advancement I have witnessed [the Human Genome Project sequenced the first human genome in 2003] has been amazing – revolutionary, even!”
Whole genome sequencing has had far reaching consequences for patient care already, from facilitating cancer stratification to improving the diagnosis of rare genetic diseases. Yet there is still plenty of progress to be made.
According to Salisbury NHS Foundation Trust, waiting times for a genomics test for a rare disease averages 84 days. As anyone anxious to hear those results will tell you, that is an excruciatingly long wait. While Nick is quick to point out the progress that’s been made (until only a few years ago, the average time to diagnosis was in excess of five years and part of the challenge remains in how hospitals engage with patients) he acknowledges that many of the issues causing delays start in the lab.
“Often, there are significant backlogs in testing, or patients testing negative are rarely retested,” he explains. “There’s simply not enough capacity and funding in the system to conduct genomic testing at the scale and quality required. Most of the time, it’s just logistically and financially impossible.”
Yet improving the efficiency, quality and speed of results will be critical as we move into a new era of genomic testing: An era which sees the true potential of genomics take off; an era where people can be diagnosed proactively, and an era where many diseases are treated or prevented long before they become fatal, or life changing.
“Improving cancer diagnosis is one of the best examples of the impact genomic testing can have,” says Nick. “Identifying the genetic signature of a particular tumour can lead to a rapid turnaround of results and significantly improve the outcomes and survival rate of a patient.”
When labs are able to sequence an individual’s genome to understand which diseases they may be at risk of, genomic testing will completely revolutionise the way we view health and medicine. Instead of having to make reactive decisions and access treatment, people will be able to use this data to make informed decisions about their own health.
“People want the most knowledge about their health possible. Genomic testing is all about gathering the data that allows patients to make informed choices. Take NIPT testing for example, knowing the conditions a baby might have can help expectant parents plan and make informed choices. In a world where we can provide genomic testing at scale, this will be possible for all families.”

While achieving the kind of efficiency required to take this step forward in patient care is a huge challenge, there have been recent advancements that have made it seem possible.
“We’ve just seen the amazing success of high-throughput, population scale COVID testing,” says Nick. “The Lighthouse Laboratory at Alderley Park managed to process over 20 million COVID tests, proving that it is possible to set up facilities and test huge volumes of samples in a relatively short space of time; this was possible mainly because of the amount of time and investment they put into process and automation.”
Automation is something that can be incredibly effective when it is applied strategically in a lab, argues Nick.
“The Lighthouse Labs show us the impact that a good, automated lab with standard operating procedures can have on improving throughput, ultimately for the benefit of patients. For automation to be effective, it’s really about achieving economies of scale, improving quality and consistency, while reducing the need for manual intervention. Routine activities such as sample preparation and aliquoting take up a lot of a lab scientist’s time; the concentration required to manually process thousands of samples accurately becomes a real issue along with the physical impact such as repetitive strain injury (RSI). Automation is required to minimise the risk of error and to provide consistency. In this way, automation can be used to streamline everything from sample receipt to analysis, so FTE time can be used more effectively.”
This ability to automate whole workflows, even entire labs, is very much a possibility today. Yet, while this kind of end-to-end automation exists, most pathology labs continue to perform much of the workload manually with a limited use of automated workflows, such as liquid handling. Meanwhile, other industries such as automotive manufacturing have long relied on fully automated facilities to great effect. We asked Nick why this level of automation is still perceived as unachievable for many labs.
“A big challenge is that while some people recognise the benefits of robots and automation, there is still a reluctance to fully embrace new technology. This can be based on factors such as concerns regarding job security – “will we be replaced by robots?”; although it may boil down to factors such as technophobia. Not everyone in the lab is sufficiently technically adept to programme/run automated systems.
“The solution is education: Demonstrating the benefits of automation is key to winning the hearts and minds of laboratory staff. The ability to improve quality, reduce and remove errors and work in a cost and time efficient manner enables the scientists to focus on what they do best – data analysis, interpretation and reporting. Ultimately, automation is there to facilitate accurate and timely generation of results – shortening the time to diagnosis and improving the management and treatment of patients.”
Even when staff are on-board, finding the right automation solution can be a challenge. Nick identifies two issues that are critical to helping automate genome sequencing: end-to-end process automation and interoperability.
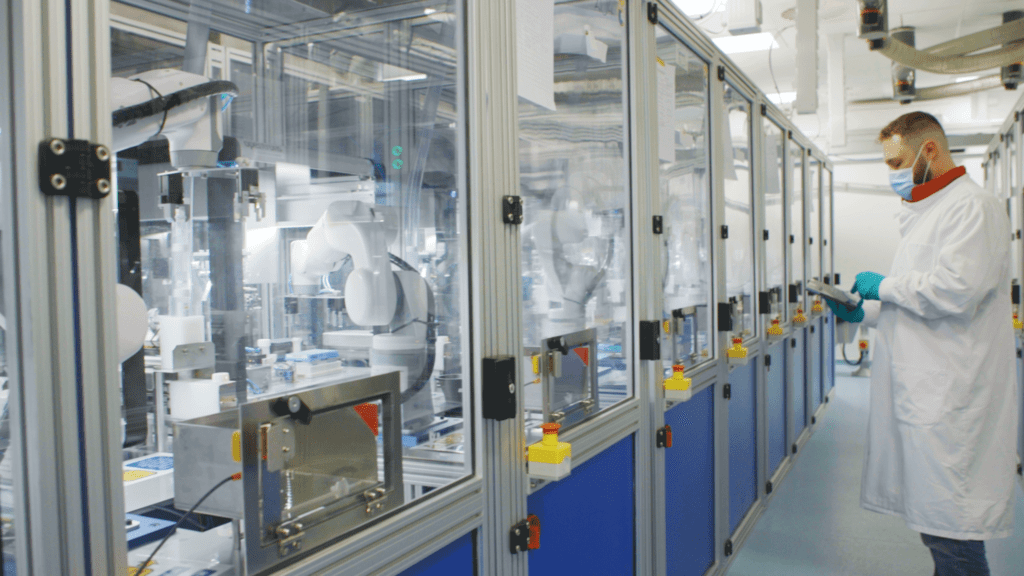
“End-to-end automation is absolutely the way to go,” enthuses Nick. “However, it can be really challenging because labs will use different equipment. That’s why interoperability is super important. There’s always going to be innovation within a space as exciting as lab automation, so solutions should be able to keep up to date and be able to work with the next new platform. Plus, people have their own favourite systems, and a loyalty to a particular brand. The automation providers who can be flexible about which systems they work with, and integrate multiple systems, will be the ones that succeed; thereby ushering in the next era of genomic sequencing.”
Learn more about Automata’s end-to-end automation solution.
