Fully Automated Solutions For Genomics
The benefits of automated solutions for Genomics
High-Throughput, Multi-Sample Compatibility
Modular and Scalable Design
True End-to-End Workflow Automation
Develop your Genomics workflow the way you want it

LINQ Canvas
Design and automate complex, multi-step genomics workflows — all without writing a single line of code. Our intuitive, color-coded labware makes experiments easy to read, edit, and plan. Reuse labware, reagents, and data flows seamlessly to build real-world genomics automation on a truly code-free platform.
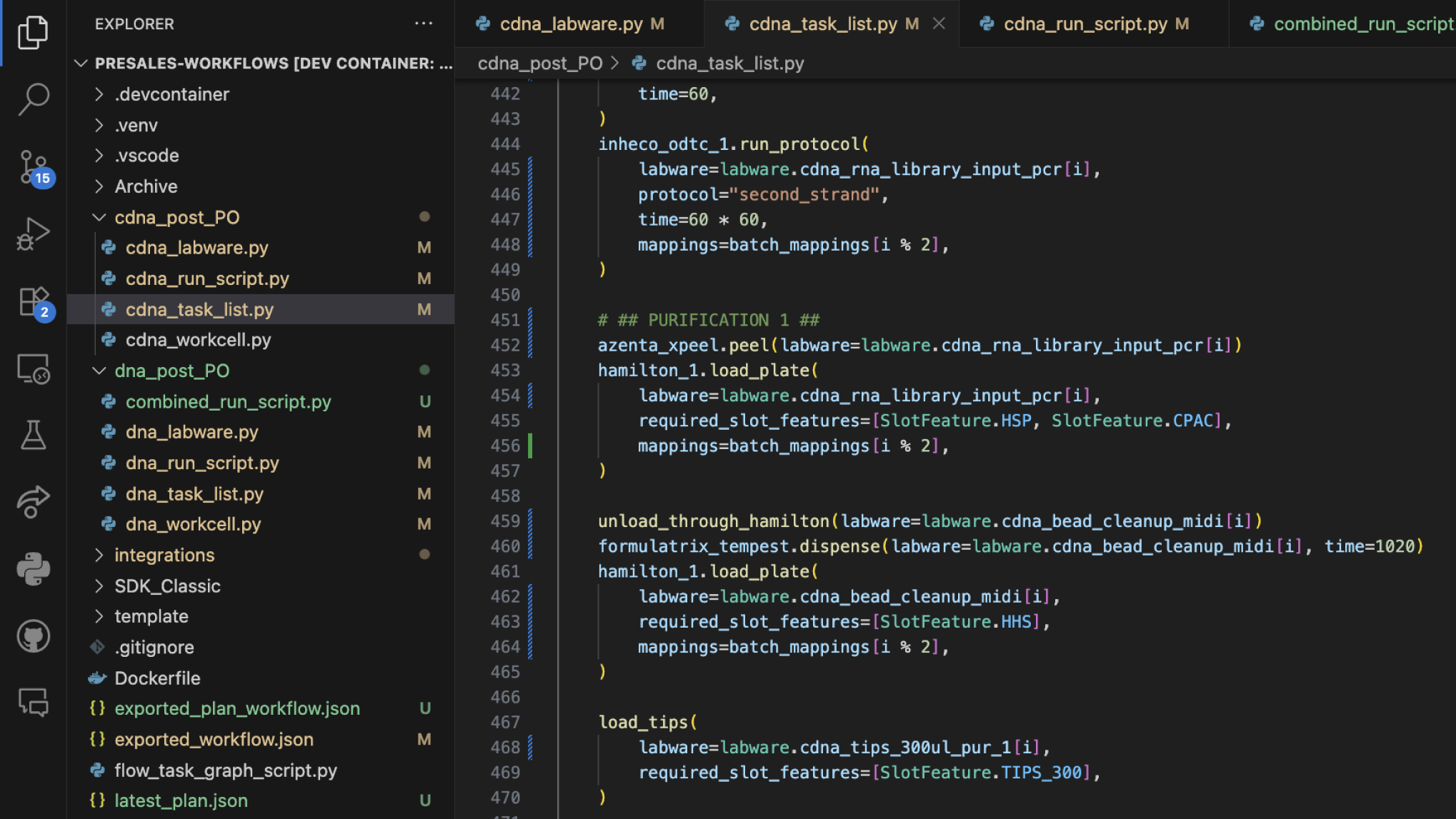
LINQ SDK and API
Take your genomics workflow building to the next level with our powerful SDK. Create custom Python functions to accelerate development, generate detailed workflow and hardware utilization metrics, and leverage custom metadata to enhance data interoperability. Seamlessly prepare your workflows for advanced machine learning and AI applications — all within a unified, intelligent platform.
Trusted by our partners












Automata partners with The Francis Crick Institute on integrated automation for Genomics
Integrated automation will support the Crick’s cutting-edge advanced genomics laboratory with greater walkaway time, R&D flexibility and data quality than ever before. By providing an integrated automated library preparation solution, together we are accelerating the pace of discoveries.
Book a meeting
Scale accurate data generation in your lab with intuitive workflow creation and data management.
Talk with our Automation experts for a personalised walkthrough of LINQ.


